Open Sourcing Deep Learning Models for Healthcare
09 Dec 2021 | deep learning healthcare opensource- The Need for Open-Source
- Deep Learning in Healthcare
- Foundational Model Use
- FDA Approved Algorithms
- The Reproducibility Crisis
- Open-Source Datasets
- Conclusion
- References
The Need for Open-Source
The recent advancements in deep learning has lead to a wide spread adoption across fields. It shows particular promise for healthcare and many companies are working towards generalizable AI for healthcare. However these models must be adopted with caution and weeded for bias similar to the recent studies done on BERT and GPT-3. Open sourcing these foundational models and future medical foundational models is key to AI safety and widespread adoption and trust of these models.
Deep Learning in Healthcare
The advancement in machine learning, particularly deep learning shows huge promise for healthcare. Deep learning has been shown to perform well with pattern recognition and problems that require utilizing massive amounts of data. With the ever increasing amount of medical data being generated (150 exabytes or 10^18 bytes in United States alone, growing 48% annually), bio-medicine stands to benefit greatly from deep learning [9]. Training of deep learning models requires access to this data, however, privacy and data sharing remain issues. There are large open-source datasets such the UK Biobank, though not on the scale of the data being generated.
Deep learning has the ability to incorporate and be adapted to all types of medical data. Recurrent Neural Networks (RNNs) and other types of Natural Language Processing (NLP) methods are heavily used for processing electronic health records (EHRs), genomics and other text based data. Convolutional Neural Networks (CNNs) can be modified for many types of medical image data. Graph Convolutional Networks (GCNs) are being used to create protein prediction networks and model drug interactions. Even physical tasks such as suturing could be learned and improved through reinforcement learning on medical robots. Novel architectures in deep learning are constantly being adapted and utilized within the healthcare research space.
The most common models utilized are created with supervised learning. These models use labeled data such as cancer type and grade with the corresponding EHRs, genomic profile, or medical image to predict known outcomes such as survival. The ability of deep learning to incorporate multimodal and temporal data also allows for novel findings. For example, perhaps a gene mutation could be shown to predict a specific tumor type shown in a biopsy image. These findings could lead to advancement in the basic medical knowledge of the disease. Utilization is limitless with diagnosis, prognosis and drug interactions just the start of what could be possible.
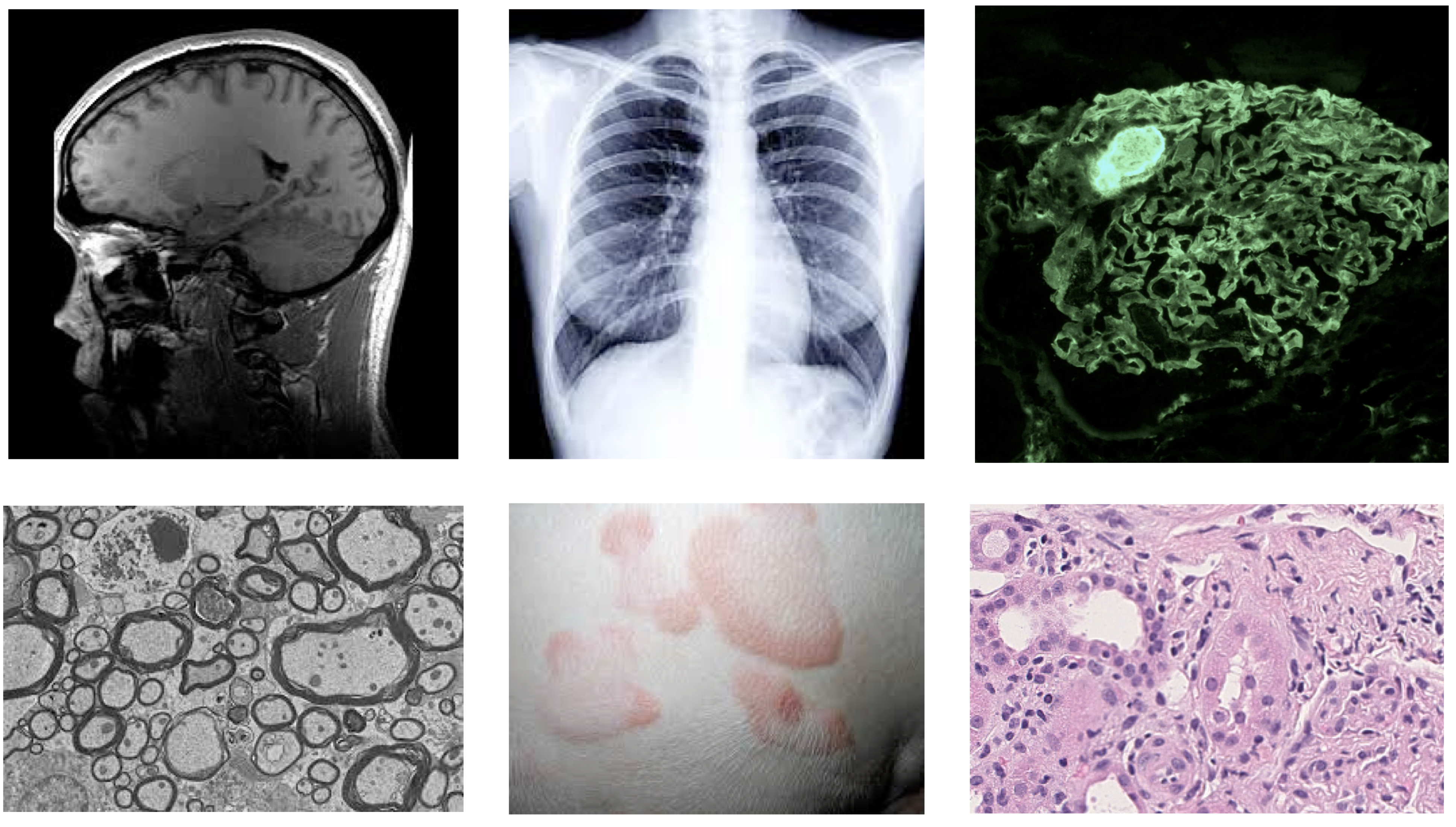
Figure 1. Various Medical Image Inputs: MRI, X-Ray, Immunofluorescence, Electron Microscopy, Dermatology, Histology
 Figure 2. Additional Medical Data Inputs: Electronic Health Record, Genomic sequencing, Protein Network
Figure 2. Additional Medical Data Inputs: Electronic Health Record, Genomic sequencing, Protein Network
Foundational Model Use
Foundational Models such as BERT and GPT-3 have the ability to adapted to almost any task. The models themselves are also being iterated upon with new architectures such as RoBERTa [10]. These models are steps towards Artificial General Intelligence. However, the models have been shown to have bias and produce violent responses [7,8]. In one instance, it told a fake patient to kill themselves [8]. These models are not currently meant to be used as medical assistance tools and creators of both models state so in their release statements. However, it is common within the cross discipline deep learning and specifically the deep learning for healthcare space to adapt architectures and models for non intended use cases. Investigating bias and other issues before this adaptation would help downstream applications later on. The models and general architecture for BERT have been released, thereby allowing independent researchers to investigate. This will also help downstream developers to investigate issues and determine if they were introduced from the base model or at a further stage.
| Foundational Model | Link | Note |
|---|---|---|
| GPT-3 | https://openai.com/blog/openai-codex/ | API is available, base code is not, small datasets available |
| BERT | https://github.com/google-research/bert | 24 Pretrained models available, some datasets available, need preprocessing |
FDA Approved Algorithms
The FDA has noted that their regulations were not designed for adaptive artificial intelligence and machine learning technologies. Their updated protocols involve extensive pre-market reviews for software modifications, incorporation of software based risk protocols and post-market monitoring to assure patient safety [5]. However, these reports are not public and companies are not required to categorize their technology as AI based.
Even with widely used algorithms, bias can arise [6]. The nature of proprietary algorithms makes analysis and investigation of bias of these algorithms difficult. Without the training data, objective function and prediction methodology the root cause of the algorithmic bias cannot be known [6]. Independent researchers are limited to an looking from the outside looking perspective, relying on work-arounds and having limited scope.
| FDA Medical AI | Link |
|---|---|
| FDA Approved Medical AI for Imaging | https://aicentral.acrdsi.org/ |
| FDA Approved Medical AI Enabled Medical Devices | https://www.fda.gov/medical-devices/software-medical-device-samd/artificial-intelligence-and-machine-learning-aiml-enabled-medical-devices |
| FDA Approved AI based Software | https://medicalfuturist.com/fda-approved-ai-based-algorithms/ |
The Reproducibility Crisis
There is an ongoing reproducibility crisis across science with over 50% of scientists in one study agreeing it is a significant crisis [1]. Biology and Medicine were the fields with the highest concerns for reproducibility [2]. This is also a concern within machine learning and a core part of why the ICLR blog track and other reproducibility conference tracks were created [3]. Machine Learning and other computationally based experiments have a relatively lower barrier to reproducibility compared to non computational experiments. Access to both the datasets and base code should be enough for replication, however, not all journals require the datasets and code for peer review and even less studies open source their datasets or code to the public [4].
Open-Source Datasets
Large, diverse datasets are needed for the advancement of deep learning for healthcare. Just as ImageNet allowed for the rapid progress in deep learning in computer vision, deep learning for healthcare also needs large, robust datasets to lower the entry barrier for researchers and to provide a baseline upon which to test models. However, due to privacy concerns it is often difficult for such data to be shared. Additionally, large datasets are hard to gather from single institutions and may not be representative of the population. This is recognized as an international problem and governments have begun to provide datasets of patients aggregated from across the country. The UK Biobank and NIH All of Us are two of the largest and comprehensive biomedical datasets that currently exist. Additionally challenges such as those on Kaggle which host datasets and code are a great boon to opensource developments. The nature of Kaggle in which an unseen, held out test set is used to compare models is a great step towards mitigating the reproducibility crisis.
| Medical Dataset | Link | Description |
|---|---|---|
| UK Biobank | https://www.ukbiobank.ac.uk/ | Extensive genetic and health information from over 500,000 patients (Note: will need approval for full access) |
| NIH All of Us | https://databrowser.researchallofus.org/ | Electronic Health Records, survey responses and other data from over 116,000 patients |
| The Cancer Genome Atlas | https://portal.gdc.cancer.gov/ | Sequencing, bio-samples, medical images from over 85,000 cases |
| 1000 Genomes Project | https://registry.opendata.aws/1000-genomes/ | Sequencing from over 2,500 patients from distinct populations |
| Deep Lesion | https://nihcc.app.box.com/v/DeepLesion | CT images from NIH, 32,000 lesions from 4000 patients |
| Human Microbiome Project | https://portal.hmpdacc.org/ | Microbiome sequencing of over 31,000 cases |
| List of other medical imaging datasets | https://github.com/sfikas/medical-imaging-datasets | |
| List of other biological datasets | https://en.wikipedia.org/wiki/List_of_biological_databases |
Conclusion
There is currently no medical AI on the scale of GPT-3 or BERT. Nonetheless we must prepare for such a future with steps towards greater transparency. However, we may never get to such an AI without reproducibility in published methods and greater availability of datasets. This would allow us to build and iterate at a greater pace and lower the barrier for investigations from independent researchers. The continued use of deep learning in healthcare makes the benefits and risks to human life and society too great to not take these measures.
References
[1] https://www.nature.com/articles/533452a
[2] https://www.nature.com/articles/s41746-019-0079-z
[3] https://neuripsconf.medium.com/designing-the-reproducibility-program-for-neurips-2020-7fcccaa5c6ad
[4] https://www.cos.io/initiatives/top-guidelines
[5] https://www.fda.gov/medical-devices/software-medical-device-samd/artificial-intelligence-and-machine-learning-software-medical-device
[6] https://www.science.org/doi/full/10.1126/science.aax2342
[7] https://aclanthology.org/W19-3823/
[8] https://artificialintelligence-news.com/2020/10/28/medical-chatbot-openai-gpt3-patient-kill-themselves/
[9] https://www.nature.com/articles/s41591-018-0316-z
[10] https://arxiv.org/abs/1907.11692